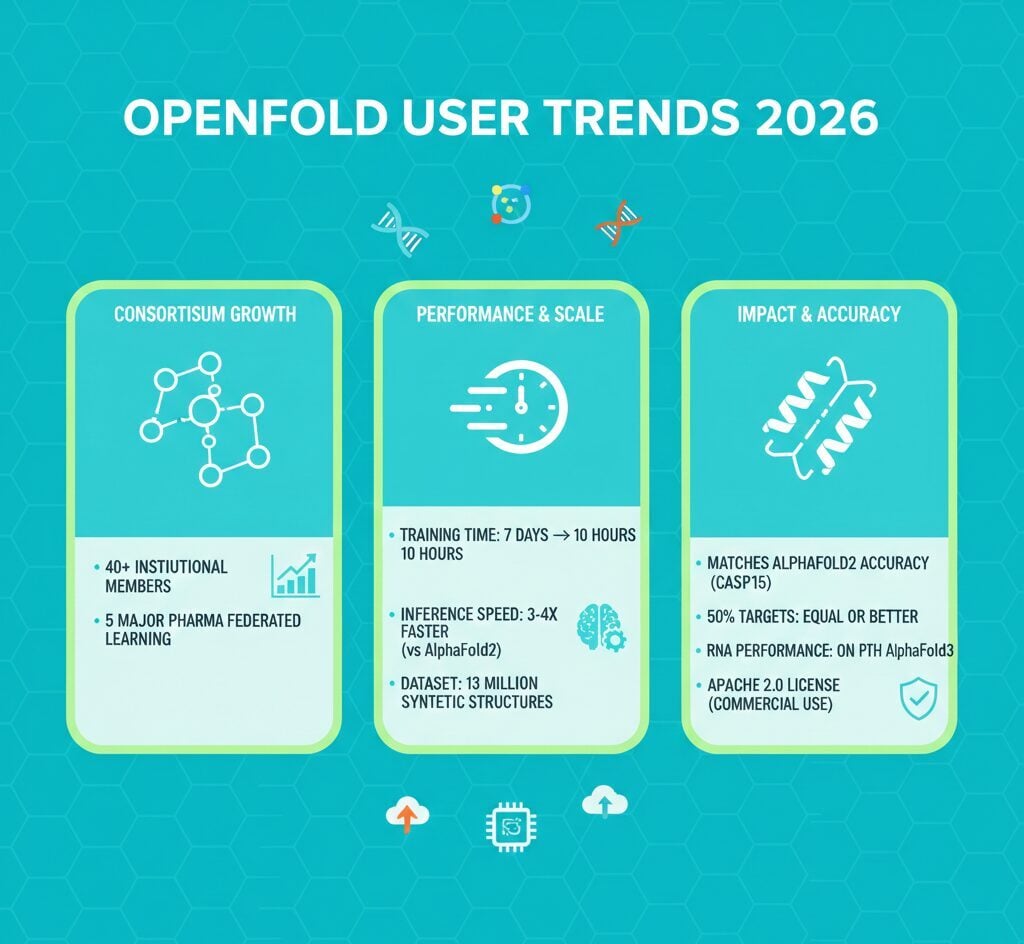
OpenFold has reached over 40 institutional members and trained on 13 million synthetic structures as of October 2025, establishing itself as the leading open-source alternative for protein structure prediction. The platform reduced training time from 7 days to 10 hours while matching AlphaFold2 accuracy on CASP15 benchmarks. The consortium now includes five major pharmaceutical companies participating in federated learning initiatives, processing approximately 20,000 protein-drug interactions from proprietary datasets.
OpenFold Key Statistics
- OpenFold consortium expanded to 40+ member institutions globally as of October 2025, including Bristol Myers Squibb, Novo Nordisk, and Johnson & Johnson
- Training dataset comprises 13 million synthetic structures, 300,000+ experimental structures, and 400,000+ multiple sequence alignments
- Inference speed reached 3-4x faster than AlphaFold2 on single A100 GPU with 13x peak memory reduction
- Training time decreased from 7 days to 10 hours using 2,080 NVIDIA H100 GPUs through ScaleFold optimization
- Five top-20 pharmaceutical companies contribute 4,000-8,000 proprietary protein-drug pairs each through federated learning
OpenFold Consortium Membership Growth
The OpenFold Consortium operates through the Open Molecular Software Foundation and recorded significant expansion throughout 2024 and 2025. The organization added 6 members in August 2024, followed by 8 members in April 2025, and 2 additional members in October 2025.
Notable organizations joining the consortium include Astex, Biogen, SandboxAQ, Psivant, Bristol Myers Squibb, Novo Nordisk, Lambda, Apheris, and Johnson & Johnson. Founding members include Columbia University’s AlQuraishi Lab, Arzeda, Cyrus Biotechnology, Outpace Bio, and Genentech’s Prescient Design.
OpenFold Training Dataset Scale
OpenFold3 utilized the most extensive training dataset among open-source co-folding models. The platform processed over 300,000 experimentally determined protein structures from the Protein Data Bank.
The OpenFold-curated synthetic structure database contains 13 million training structures. The OpenProteinSet provides 400,000+ multiple sequence alignments and template files for comprehensive model training.
Training infrastructure leveraged AWS cloud computing with 256 GPUs, delivering 85% cost savings compared to on-demand pricing through EC2 Capacity Blocks and Spot Instances. This collaborative approach involved Novo Nordisk, Columbia University, and AWS.
OpenFold Performance Improvements
OpenFold demonstrates substantial computational advantages over baseline implementations. Inference speed reached 3-4x faster than AlphaFold2 on a single A100 GPU.
The DS4Sci Kernel optimization achieved 13x peak memory reduction compared to standard implementations. Custom kernels reduced GPU memory usage by 4-5x versus FastFold and PyTorch equivalents.
Maximum sequence length processing reached 4,000+ residues on a single A100 with CPU offloading capabilities for longer chains. The ScaleFold optimization methodology reduced training time from approximately 7 days to just 10 hours.
OpenFold Benchmark Results
ScaleFold completed the OpenFold partial training task in 7.51 minutes during MLPerf HPC v3.0 benchmarks, achieving over six times the speedup compared to baseline performance.
OpenFold Accuracy Statistics
OpenFold matched established gold standards across multiple evaluation metrics. The platform achieved competitive performance on CASP15 with a GDT-TS 95% confidence interval of 68.6-78.8, compared to AlphaFold2’s range of 69.7-79.2.
| Benchmark | OpenFold Result | Comparison |
|---|---|---|
| CASP15 GDT-TS | 95% CI: 68.6-78.8 | AlphaFold2: 69.7-79.2 |
| Target Parity | 50% of targets | Equal or better than AlphaFold2 |
| RNA Performance | Matches AlphaFold3 | Only open-source at parity |
| Protein-Ligand Co-folding | On par with AlphaFold3 | Outperforms Boltz-2 |
OpenFold3 became the only open-source model to match AlphaFold3’s performance on monomeric RNA structures. The model shows robust generalization capacity, achieving high accuracy even when training set size reduced to 1,000 protein chains.
OpenFold Federated Learning Initiative
The AI Structural Biology Network coordinates federated learning initiatives through Apheris, enabling OpenFold3 training on proprietary pharmaceutical data while protecting intellectual property. Five top-20 pharmaceutical companies participate, including AbbVie, Astex, Bristol Myers Squibb, Johnson & Johnson, and Takeda.
Each participating company contributes 4,000-8,000 proprietary protein-drug pairs. The aggregated federated model accessed approximately 20,000 protein-drug interactions.
The federated approach enables training on approximately five times more proprietary structural data relevant to industrial drug discovery compared to the Protein Data Bank alone. Public databases contain only about 2% of drug-like structures.
OpenFold Licensing Comparison
OpenFold operates under Apache 2.0 licensing, distinguishing it from proprietary alternatives. The platform permits unrestricted commercial and academic use, contrasting with AlphaFold3’s non-commercial restrictions.
| Feature | OpenFold | AlphaFold3 |
|---|---|---|
| License Type | Apache 2.0 | Non-commercial only |
| Commercial Use | Yes | No |
| Training Code | Available | Not available |
| Fine-tuning | Permitted | Not permitted |
| Classification | Class 1 (Linux Foundation) | Proprietary |
OpenFold3 accessibility includes GitHub repository code, Hugging Face model checkpoints and Docker images, Tamarind Bio’s hosted interface, and Apheris’ locally deployable versions. NVIDIA NIM integration provides containerized, accelerated API deployment for scalable inference workflows.
OpenFold Industry Adoption
Major pharmaceutical and biotechnology companies integrated OpenFold into research pipelines. Novo Nordisk utilizes the platform for internal discovery pipelines with proprietary data integration.
| Organization | Application Focus | Use Case |
|---|---|---|
| Novo Nordisk | Internal Discovery | Proprietary data integration |
| Bayer Crop Science | Agricultural Research | Plant, weed, pest proteins |
| Outpace Bio | Cell Therapy | Molecular circuits design |
| Cyrus Biotechnology | Enzyme Therapeutics | Autoimmune treatments |
| Meta AI | Protein Atlas | 600M+ protein characterization |
Meta AI utilized OpenFold to launch an atlas featuring over 600 million proteins from bacteria, viruses, and other microorganisms. The UK government’s OpenBind initiative committed to refining OpenFold3 using collaborator-generated data.
OpenFold Computing Infrastructure
Development and deployment rely on substantial computing infrastructure across multiple supercomputing centers. The Texas Advanced Computing Center provided Frontera and Lonestar6 supercomputers for large-scale machine learning deployments.
AWS contributed EC2 Capacity Blocks and Spot Instances with 256 GPUs for OpenFold3 training. The NVIDIA Eos Supercomputer utilized 10,752 H100 GPUs for ScaleFold optimization benchmarks.
According to Dr. Nazim Bouatta at Harvard Medical School, supercomputers combined with AI enable prediction of 100 million structures in just a few months. The Texas Advanced Computing Center provided crucial allocations for breakthrough performance.
FAQ
How many institutions are members of the OpenFold Consortium?
The OpenFold Consortium comprises over 40 member institutions globally as of October 2025, including pharmaceutical companies, biotechnology firms, academic laboratories, and technology companies.
What is the size of OpenFold’s training dataset?
OpenFold3 trained on 13 million synthetic structures, 300,000+ experimentally determined protein structures from PDB, and 400,000+ multiple sequence alignments from OpenProteinSet.
How fast is OpenFold compared to AlphaFold2?
OpenFold achieves 3-4x faster inference speed than AlphaFold2 on a single A100 GPU. Training time reduced from 7 days to 10 hours using ScaleFold optimization.
What license does OpenFold use?
OpenFold operates under Apache 2.0 licensing, permitting unrestricted commercial and academic use. This contrasts with AlphaFold3’s non-commercial only license restrictions.
How many pharmaceutical companies participate in federated learning?
Five top-20 pharmaceutical companies participate in the federated learning initiative: AbbVie, Astex Pharmaceuticals, Bristol Myers Squibb, Johnson & Johnson, and Takeda.